|
|
Perspective
An Open Innovation Ecosystem for Drug Discovery
By Michael R. Barnes, PhD
Director of Bioinformatics, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, John Vane Science Centre, Charterhouse Square, London, EC1M 6BQ, UK
2OI Pharma Partners, Red Sky House, Fairclough Hall, Halls Green, Weston, Hertfordshire, SG4 7DP.
dr.michael.r.barnes@gmail.com
Survival of the fittest
Few would disagree that the Pharmaceutical industry has been experiencing a rather severe productivity crisis in the last several years. As a Computational Biologist working in the Industry and now transitioning to academia, it has been interesting to experience this crisis both at first hand and vicariously through the experiences of colleagues and peers in other companies. On a personal level, this has helped me put many of the commentaries on pharma woes, which range from blatant iconoclasm to dogmatic denial, into a wider perspective. A common theme that emerges is that the industry is experiencing unprecedented changes. Naturally all perspectives are shaped by personal experience, but my own experience suggests that the current crisis in the pharmaceutical industry stems from a failure to evolve in response to these changes, as market conditions and the expectations of pharma have shifted seamlessly from largely favourable to quite hostile. The situation seems entirely Darwinian in nature. As environments change, those which are able to adapt - the fittest, survive. Successful strategies clearly exist for companies, for example, shifts away from small molecule drug discovery towards biopharmaceuticals, have seemed successful at least in the short to medium term for some companies, Roche being a good case in point1. Other companies, such as GlaxoSmithKline, have also tried to adopt the biotech operating model with the creation of 38 internal discovery performance units (DPUs), funded on three year cycles by an internal investment board, designed to emulate the venture capital funding process2. These trends are mirrored to variable extents across the sector, but to date most changes have been largely tactical, underpinned by merger and acquisition, rather than a step change in the capacity to innovate.
This perspective will try to provide some suggestions to improve the innovative capacity of the drug discovery process, particularly including chemistry, by increasing partnership and data sharing based on the principles of open innovation. Ultimately, I propose that the interdependencies between the various players of the drug discovery process are much greater than may have been anticipated. Taking a biological view of the problem, I suggest that the fates of the leading protagonists in the “innovation ecosystem” are intricately interconnected with the smallest players.
On the Origin of Open Innovation
The term Open Innovation was coined by Henry Chesbrough as recently in 20033, but in reality the principles of open innovation were established much earlier. Arguably open innovation goes back a long way, even if not by name. Some have even pointed as far back as the Medici dynasty of 16th Century Florence, whose patronage of the Arts and Sciences led to some of the multidisciplinary advances that fuelled the Renaissance4. But debating the origins of the term open innovation probably distracts from the simplicity of the concept. Open innovation simply argues that great ideas can come from anywhere and should be able to go anywhere. Importantly it also maintains that fair rights of idea ownership should be retained, but should not be a barrier to the movement of these ideas. The later is perhaps the key concept that differentiates open innovation from other forms of innovation. Ideas should not be monopolised by one individual or organisation. This does not mean that intellectual property is at odds with open innovation, perhaps the only concept at odds with open innovation is exclusivity. As soon as an idea is exclusively owned and controlled its potential to evolve is greatly diminished. Going back to the ecosystem analogy, it is in effect removed from the gene pool.
Why Open Innovation is not an easy option
Open innovation has been widely recommended as a fix-all solution to poor productivity in pharma, but there are a few fallacies about the concept that should be considered, as they could potentially lead to a future backlash against the concept. The first is that an open innovation strategy can be implemented exclusively by specialists in legal, IP and business development. Although there is clearly an important role for such specialists in seeking innovation, negotiations and deal making, arguably many of the failings of open innovation to date have been related to the failure to involve “hands on” internal domain experts in the evaluation of external technologies. A very recent example, being the relative failure of GSK to translate compounds and tools acquired from the biotech company, Sirtris5. This seems to be a trend in Pharma, where the critical mass of disease or technological expertise, in the traditional “therapeutic area” has been dispersed in favour of opportunistic externally facing drug discovery units and venture units. The success of this concept relies heavily on the excellence of the staff leading the external facing units and perhaps uncovers a paradox of the open innovation concept. That is that the best Innovators tend to be very active (“hands on”) in their field, but an open innovation strategy, by focusing externally rather than internally, may not favour “hands on” involvement in the innovation process. This argues for a very tight partnership between specialist “innovation seekers” and the internal innovators (read ordinary lab scientists) who can ensure the translation of external ideas into internal successes. So it follows therefore, that open innovation has the best potential to work within the pre-existing R&D framework of organisations. The emphasis here is on organic growth and cultural change within organisations, rather than radical solutions and re-structuring. Success of this strategy is in many cases dependent on the visibility of “external innovations” within and across fields, so that they can be recognised and adopted. This is probably the biggest challenge for a successful open innovation strategy.
Publicise or die...
Although the traditional patent document is the conventional route to describe and protect innovation, without other publicity strategies it can be a serious barrier to open innovation. This is not in terms of the intellectual property claimed, but due to the tendency of patent documents to obfuscate claims, either intentionally or unintentionally6. Publication is the most widely used mechanism for publicising innovation, usually after intellectual property (IP) has been secured. But a publication focused strategy brings with it real problems for innovation seekers. Foremost among these is the peer review process itself, which can introduce multiple biases into the published literature corpus7. For example, the so-called “Matthew effect” has been well documented, showing that reviewers and editors tend to be much more favourable in their evaluation of manuscripts submitted by “famous investigators” from prestigious institutions, regardless of the manuscripts’ scientific and technical merit7. The high impact and palpable excitement in the literature surrounding resveratrol8, undoubtedly played a role in the acquisition of Sirtris for $720 million. The issues with peer review also make it comparatively much more difficult for new investigators, or investigators in new fields to publish in high impact journals. Also at a more fundamental level, some really great innovations may not be considered worthy or substantial enough to support high impact publication alone. Instead they need to be recognised and adopted within the wider translational research process to show their true value.
Does Crowd Science have a place in drug discovery?
Crowd Science is a key open innovation concept to improve the visibility of the needs of innovators and innovation seekers. The concept has been implemented in many different ways, two key implementations of crowd science are considered here. From the point of view of innovation seekers, “crowd sourcing” is a process where tasks traditionally performed by specific individuals are opened up to a group of people or community (crowd) through an open call. Research funding agencies are already working in this way when they release a call for proposals in a specific research area. The other concept to consider from the point of view of those wishing to publicise their skills or innovations, is “crowd funding”. This describes an open call process to collectively support specific efforts initiated by other people or organisations.
Both approaches have already been applied to aspects of drug discovery with some success. A good example of the crowd funding approach is DrugDev.org (Table 1), which uses social networking technology to publicise the availability of over 60,000 clinical trial investigators in 93 countries. The database includes a capability to provide feedback on investigators' trial recruitment capabilities, infrastructure and quality. In less than 2-years DrugDev.org has grown from a start-up to the biggest network of independently rated research sites in the world, transforming the way many major CROs and pharmaceutical companies conduct study feasibility, site identification and startup activities, with quite a dramatic effect on timelines and cost. A well tailored Crowd funding approach to publicise translational researchers and their innovations, does not yet exist, but could be an interesting tool to improve and strengthen the interface between industry, academia and the clinic. A quick look at the success of the approach in other areas (e.g. funding of creative projects at www.kickstarter.com) highlights the potential of this approach.
Crowd sourcing is probably the most successful open innovation concept to date, the poster child being InnoCentive (Table 1), which was developed at Eli Lilly to use the internet as a route to discover solutions to challenging internal research problems. InnoCentive, became the first global Internet-based platform designed to help connect Seekers, those who had difficult research problems, with Solvers, those who came up with creative solutions to these problems. The crowd sourcing concept has now been widely adopted throughout healthcare by diverse companies including GE Healthcare, Johnson & Johnson and Procter & Gamble. The U.S. Patent and Trademark Office have also notably applied crowd sourcing to the patent review process with their Peer to Patent Community Patent Review project (Table 1), which allows scientists to submit prior art which might invalidate a patent application. Perhaps the last bastion of translational research where crowd sourcing could make a real impact would be as an alternative to the peer review process. This has been widely discussed and strongly advocated by some, but ultimately would require a huge cultural change that the translational research community may not yet be ready for.
Finally another example of the power of the crowd comes from patients themselves and shows how social media is already influencing drug discovery. Patientslikeme (Table 1) is an online social networking forum that allows patients to share treatment experiences. When a small Italian study reported that lithium carbonate had the potential to slow the progress of Amyotrophic lateral sclerosis (ALS), hundreds of Patientslikeme users started taking the drug under the supervision of their physicians9. They were unable to replicate the promising findings of the preliminary study, but nevertheless the power of sharing data to rapidly advance medicine was clearly demonstrated.
A Brave New World
Organisational and regulatory cultures can be a major contributor to the failure of open innovation in any organisation. Put simply it is not adequate to ask employees to think differently and challenge the status quo, while continuing to work entirely within the status quo, using existing tools and policies. Innovation requires new thinking and importantly the ability and permission to use new tools. A recurring issue within drug discovery is the fear of inadvertent disclosure of information. For example, some companies have a placed a moratorium on the use of public domain tools and databases, based on a largely unfounded fear of publicly disclosing proprietary sequence or small molecule information, and thus invalidating future IP claims (no precedent exists for such disclosures). The necessary requirement for patient confidentiality can also be a real barrier for innovation. This is particularly evident in the field of genetics, where large-scale genetic data sets are frequently shared with other research groups and often released into the public domain to allow for meta-analysis. Study participants are usually not informed about such data sharing because data are assumed to be anonymous after stripping off personal identifiers. However a study by Homer et al10 showed that the assumption of anonymity of genetic data is tenuous as even summary information (in the case of this study, p-values without genotype information) can be intrinsically self-identifying. This publication led to a wholesale worldwide change in data sharing policies for genetic data, severely limiting the access of non-authorised researchers to genetic data11. The implications of these rulings have probably not been fully understood as they could potentially limit open access to all datasets generated with next generation sequencing technology, severely impeding access to most of the exponentially expanding genetics and genomics corpus in the public domain. There are no simple answers to this issue, but it clearly illustrates how retaining the status quo for data access policies could substantially impede innovation to the detriment of the very patients that policies are intended to protect.
Partnering to cross the “Translational Valley of Death”
The Translational “valley of death” is a widely used concept referring to the widening gap between advances in basic science and the practical application of that knowledge into the clinic12. Historically public domain resources for drug discovery research, particularly medicinal chemistry, may have exacerbated this situation and until recently have been under-resourced and sometimes poorly curated. However this situation is rapidly changing. Governments and other science funding organisations have substantially increased translational research funding to facilitate access to both data and screening facilities. Examples of such investment include the NIH molecule libraries initiative, PubChem and the Wellcome Trust funded Chembl database (Table 1). Encouragingly a recent (crowd sourced) appraisal of the quality of the data was very positive13. This trend looks set to continue. The community impact of the increased focus on good public domain data and resources is palpable, with the announcement of major open access drug discovery projects, such as Arch2POCM (Table 1), which aims to take small molecules for Autism, Schizophrenia and Cancer into man.
Pharmaceutical companies are now following this trend by pro-actively engaging in precompetitive data sharing on an ad hoc basis14 and collectively though major public-private partnerships like the €2bn EU Innovative Medicines Initiative (IMI)15. The IMI is spawning a number of excellent translational resources, including the Open Pharmacological Space, a raft of drug discovery tools currently being constructed by the Open PHACTS consortium (Table 1). Collectively these initiatives are resulting in unprecedented access to data, information and knowledge, giving capabilities to public domain scientists that were previously only available behind industry firewalls.
Before euphoria takes hold, it’s worth pointing out that there is still a long way to go before public domain drug discovery is truly enabled. A key determinant will be the standardisation of data exchange allowing greater integration between public and private domains. The Pistoia Alliance (Table 1) was established to address this issue and appears to be making some headway. The MIABE (Minimum Information About a Bioactive Entity) initiative is also making valuable progress16. Despite movement in the right direction there is still a widespread lack of agreed standards and vocabularies that unambiguously identify the entities, processes and observations within experimental data relevant to drug discovery17. The consequences of not agreeing such standards are still evident in many of the other existing systems concerning bioactivity data which are still very difficult to exploit in a structured manner.
A new drug discovery ecosystem must evolve to “do more with less”
“Do more with less” has become the standard refrain of CEOs in most industries, with the pharma industry being no exception18. The open innovation approach to drug discovery is well placed to meet this demand, but is reliant on the dynamics of the entire drug discovery community. Now that the gap between industry and public domain drug discovery appears to be narrowing, open innovation is a natural and intuitive response to build stronger public-private partnerships. In times of austerity, considering industry woes in this area, it might seem folly for governments and funders to increase their emphasis on drug discovery, but this is happening nevertheless and may change the entire dynamics of the drug discovery community. While the US NIH seeks to speed translation with their aggressively open access strategy at the National Center for Advancing Translational Sciences19, the EU Innovative Medicines Initiative is also setting a new high water mark for pre-competitive and public-private collaborations15. Considering the trends that are affecting both public and private sectors the time has never been better for open innovation initiatives (Box 1). In some cases open innovation and open access strategies may represent the only hope for disease areas with significantly higher attrition rates compared to the norm, such as the psychiatric disease area20. Despite considerable societal need, many large pharmaceutical firms have now ceased all R&D programmes in psychiatry, because the cost of failure is perceived to be too high20. Open access or open innovation approaches could lower the barriers of entry to this area and spread risk, enticing firms to re-engage with areas of severe unmet medical need. Risk is also a relative concept. To the academic the “risk” of an unknown or complex disease mechanism is synonymous with breakthroughs and high impact publication. In essence academics are funded to address this risk and a completely negative outcome for academic research is rare. For Industry outcomes of drug discovery are starker and the financial stakes are much higher.
| Industry trends |
Academia / Public Domain trends |
- Declining budgets
- Declining productivity
- Risk aversion
(seek risk sharing)
- R&D externalisation trend
- Late stage focus
- Driver: Shareholder value
|
- Austerity measures
- Translational imperative
- Risk = Impactful publication
- Increasingly entrepreneurial
- Early & Clinical stage focus
- Drivers: Publication & Public Health
|
Box 1.
Sector trends show why public-private partnership makes sense for drug discovery
Innovation as food
Viewing open innovation from the point of view of a biologist, there are striking similarities between the innovation communities and the ecological concept of the food chain21. Extending the analogy of innovation as “food” in the innovation ecosystem, different players are clearly occupying different trophic levels (Figure 1). We could define the academics engaged in pure research as primary producers; academics engaging in applied research could be termed primary innovators; industry and SMEs at the translational interface could be termed secondary innovators; while the externally facing tech-transfer and venture units in pharma and large SME could be termed tertiary innovators. The concept fits on multiple levels. Firstly, in ecosystems about 10 % of the energy transferred between each trophic level is converted to biomass, the same might be said about the translation of ideas. Secondly, it clearly illustrates the inter-dependency of each player and the need for a strong academic sector (including pure “blue skies” research) to act as a robust foundational layer for healthy industrial sectors.
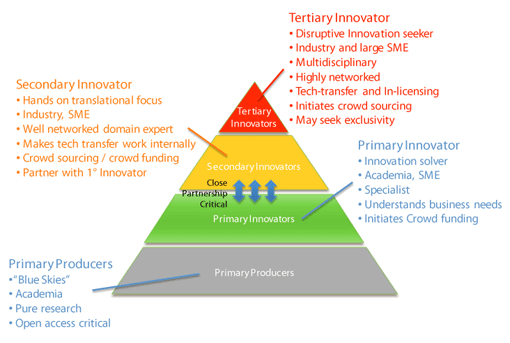
Figure 1.
An Open Innovation Ecosystem for Drug Discovery
Open Innovation or Open Access?
This perspective has conscientiously avoided distinguishing open innovation from the open access approach to drug discovery which is generating some controversy. It really remains to be seen how successful a pure open access approach will be in translating clinical candidates to market without a clear model to generate a return on investment. Clearly open access drug discovery has potentially huge societal value, but equally the patents that protect the assets of the pharmaceutical industry also have important societal value, by sustaining a vital industry. Currently there are few viable alternatives to the patent system to protect investment. Open innovation does not challenge this view, and should be largely agnostic to the approach used to define ownership of innovation, be it open access or patent protected. It is probably more accurate to view the process of innovation as a continuum from open access to secure intellectual property. Perhaps the greatest challenge to successful open innovation is the accurate determination of where the pre-competitive and competitive boundaries of drug discovery lie, chemists will play a leading role in this debate.
_________________________________________________________________
References
- Hayden EC. Roche vows to keep Genentech culture. Nature, 2009, 458, 270.
- Whalen, J. Glaxo tries biotech model to spur drug innovations. Wall Street J, 2010, July 01.
- Chesbrough H. Open Innovation: The New Imperative for Creating and Profiting from Technology, Harvard Business School Press, Boston, MA, 2003.
- Johansson, F. The Medici Effect: Breakthrough Insights at the Intersection of Ideas, Concepts and Cultures, Harvard Business School Press, Boston, MA, 2004.
- Schmidt C. GSK/Sirtris compounds dogged by assay artifacts. Nat Biotechnol. 2010, 28(3):185-6.
- Griffin TD, Boyer SK, Councill IG. Annotating patents with Medline MeSH codes via citation mapping. Adv Exp Med Biol. 2010, 680, 737-44.
- Hojat M, Gonnella JS, Caelleigh AS. Impartial judgment by the "gatekeepers" of science: fallibility and accountability in the peer review process. Adv Health Sci Educ Theory Pract. 2003, 8(1), 75-96.
- Hall SS. Longevity research. In vino vitalis? Compounds activate life-extending genes. Science. 2003, 301(5637), 1165.
- Wicks P, Vaughan TE, Massagli MP, Heywood J. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol. 2011, 29, 411-4.
- Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008, 4(8):e1000167.
- Wjst M. Caught you: threats to confidentiality due to the public release of large-scale genetic data sets. BMC Med Ethics. 2010, 11:21.
- Coller BS, Califf RM. Traversing the valley of death: a guide to assessing prospects for translational success. Sci Transl Med. 2009, 1(10):10cm9.
- Oprea TI, Bologa CG, Boyer S, Curpan RF, Glen RC, Hopkins AL, Lipinski CA, Marshall GR, Martin YC, Ostopovici-Halip L, Rishton G, Ursu O, Vaz RJ, Waller C, Waldmann H, Sklar LA. A crowdsourcing evaluation of the NIH chemical probes. Nat Chem Biol. 2009, 5(7):441-7.
- Barnes MR, Harland L, Foord SM, Hall MD, Dix I, Thomas S, Williams-Jones BI, Brouwer CR. Lowering industry firewalls: pre-competitive informatics initiatives in drug discovery. Nat Rev Drug Discov. 2009, 8(9):701-8.
- Goldman M. Reflections on the Innovative Medicines Initiative. Nat Rev Drug Discov. 2011, 10(5):321-2.
- Orchard S, Al-Lazikani B, Bryant S, Clark D, Calder E, Dix I, Engkvist O, Forster M, Gaulton A, Gilson M, Glen R, Grigorov M, Hammond-Kosack K, Harland L, Hopkins A, Larminie C, Lynch N, Mann RK, Murray-Rust P, Lo Piparo E, Southan C, Steinbeck C, Wishart D, Hermjakob H, Overington J, Thornton J. Minimum information about a bioactive entity (MIABE). Nat Rev Drug Discov. 2011, 10(9):661-9
- Harland L, Larminie C, Sansone SA, Popa S, Marshall MS, Braxenthaler M, Cantor M, Filsell W, Forster MJ, Huang E, Matern A, Musen M, Saric J, Slater T, Wilson J, Lynch N, Wise J, Dix I. Empowering industrial research with shared biomedical vocabularies. Drug Discov Today. 2011, 16(21-22):940-7.
- Witty, A. Research and Develop. The Economist, 2010, Nov 22nd.
- Collins, FS. Reengineering Translational Science: The Time Is Right. Sci. Transl. Med, 2011, 3, 90cm17
- Wong EH, Yocca F, Smith MA, Lee CM. Challenges and opportunities for drug discovery in psychiatric disorders: the drug hunters' perspective. Int J Neuropsychopharmacol. 2010, 13, 1269-84
- http://en.wikipedia.org/wiki/Trophic_level
|
|

Editor
Gabriele Costantino
Univ. of Parma, IT
Editorial Committee
Erden Banoglu
Gazi Univ., TR
Lucija Peterlin Masic
Univ. of Ljubljana, SLO
Leonardo Scapozza
Univ. of Geneve, CH
Wolfgang Sippl
Univ. Halle-Wittenberg, DE
Sarah Skerratt
Pfizer, Sandwich, UK

Executive Committee
Hans-Ulrich Stilz President
Gerhard F. Ecker Past Pres.
Koen Augustyns Secretary
Rasmus P. Clausen Treasurer
Hein Coolen Member
Gabriele Costantino Member
Phil Jones Member

|






